Do you think that integrative research has added-value? Is it more than the sum of its parts? How can we show how integrative is a body of research, and investigate if it has added-value?
These are difficult questions that we don’t have all the answers to. Here we give a few possibilities based on the corpus of publications produced by a research programme aimed at creating added-value via promoting integrative research. We visualise characteristics of that corpus of publications, and compare to a reference corpus of publications arising from research that may or may not have included integrative research. (And the gory details of how we did this are provided below.)
The example below involves the corpus of publications arising from the University of Zurich Research Priority Programme in Global Change and Biodiversity (URPP GCB). The reference corpus is from related research fields for similar time period at University of Zurich (exact specification in the gory details section).
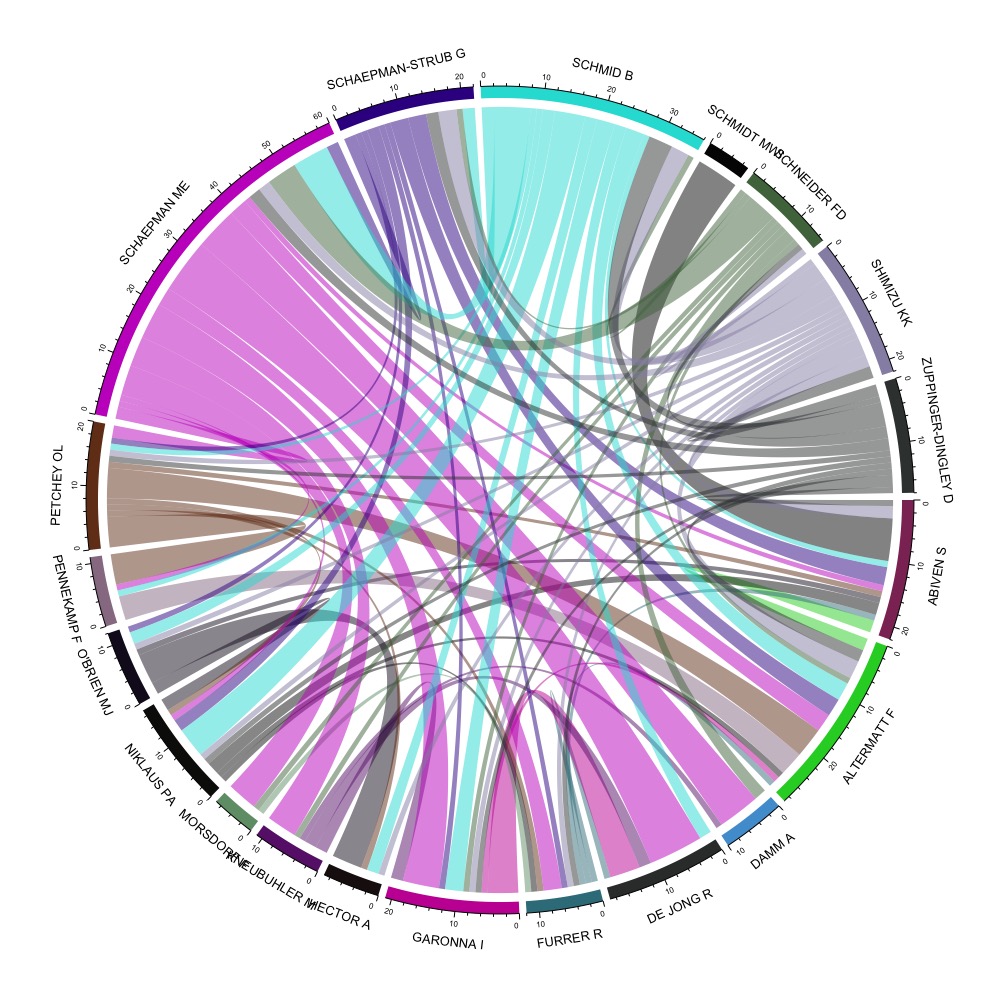
The network of co-authors in the URPP GCB corpus.
Figure 1 shows the tightly connected co-authorship network of 20 of the researchers in the URPP GCB. It shows that in terms of collaboration there is a great deal of integration. The number of co-authors in the URPP GCB corpus is about 20% greater than in the reference, though only by about one author on average (Figure 2). The number of times articles were cited is about 30% greater in the URPP GCB corpus than in the reference (Figure 2).
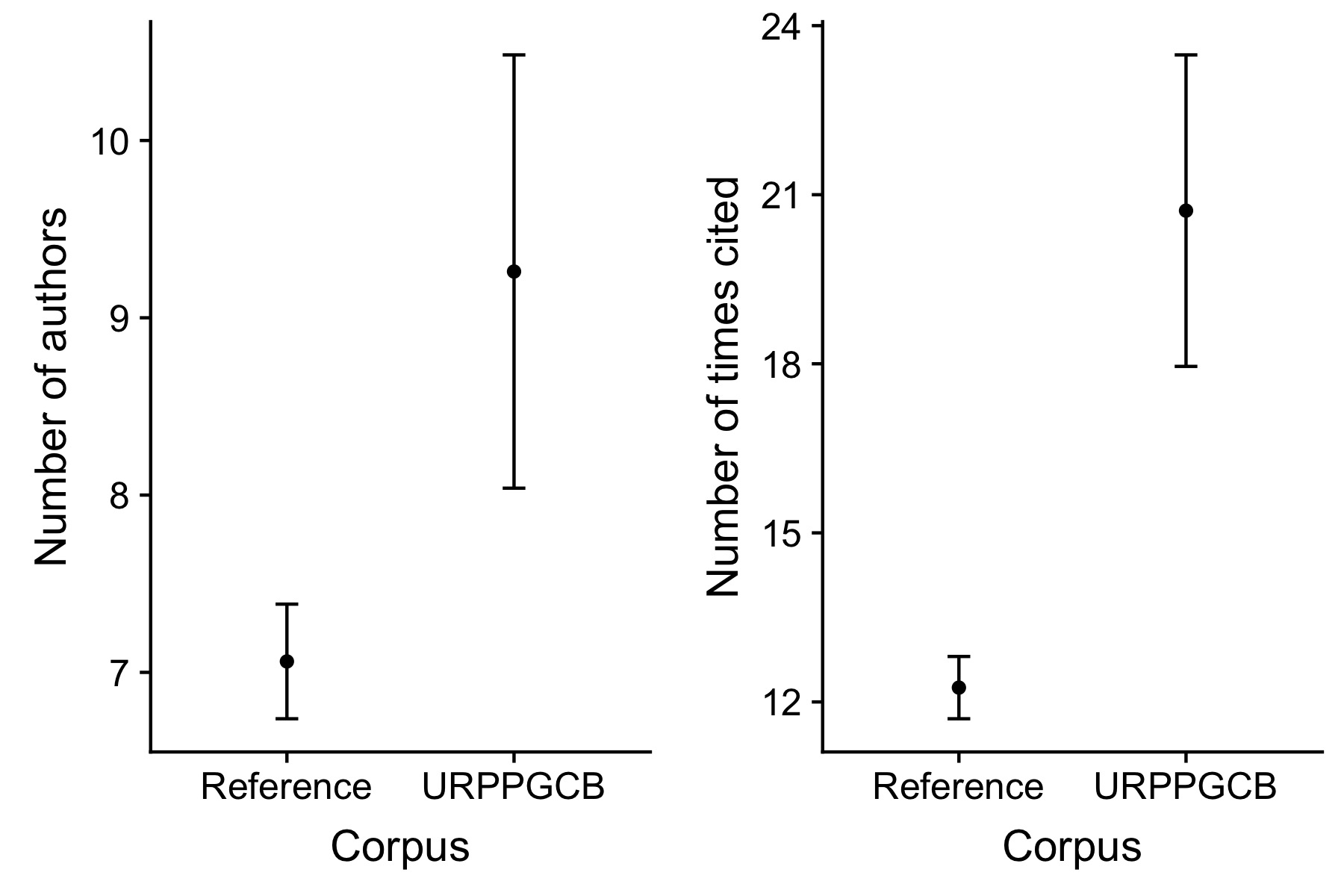
Comparison of number of authors per article and number of times articles are cited between in the reference corpus and the focal (URPP GCB) corpus. (Mean plus/minus one standard error.)
The gory details…
Warning! — what you see below is a bit technical, not all explained in great detail, and you need to know how to use R / RStudio and various add on packages.
The code and datasets used below are available in this github folder.
Assemble your focal corpus
We manually entered all publications produced by the URPP GCB into Zotero. (Actually, we used the lovely Add items by identifier Zotero button, and added by DOI.)
Get information about your corpus from WoS
We will get information from WoS on the specific papers in our corpus by searching WoS for the DOIs of the articles in the corpus. To do this, we want to get text, which we’ll paste into the WoS search term field, of the following nature :
02/ECY.2003 OR 16/J.ECOCOM.2016.12.005 OR 16/J.RSE.2014.11.014 OR 02/ECY.1748 OR 16/J.COSUST.2018.04.016 OR 02/ECE3.1186 OR 93/FEMSEC/FIV066 OR 11/ELE.12443
To get this text:
## install the following if required
library(bibliometrix)
library(tidyverse)
library(circlize)
library(wordcloud2)
library(cowplot)
- Highlight the papers in Zotero that are your corpus.
- Use the File -> Export Library… tool in Zotero, choose BibTex format, leave all boxes unchecked, leave Character encoding as Unicode (UTF-8), click OK, and choose the location and filename. We named the file
from_zotero.biband saved it to our Desktop (referred to on a Mac by~/Desktop/). - Use the following code in R to get a new file that contains the text, mentioned above, that we will paste into the WoS search:
readFiles("~/Desktop/from_zotero.bib") %>%
isibib2df() %>%
dplyr::mutate(DOI=stringr::str_trim(DI)) %>%
filter(!is.na(DOI)) %>%
pull(DOI) %>%
paste(collapse=" OR ") %>%
cat(file="~/Desktop/dois_for_WoS.txt")
- Open the file (we named it
dois_for_WoS.txtand saved it to our Desktop) with the DOI text we need to paste into the Wos search field. Copy the text, and paste it into the search term window of WoS Basic Search. SelectDOIas the item being searched for. Hit the Search button. - You may get fewer results from WoS than DOIs you entered. At time of writing this article we had 164 DOIs, or which WoS found 138. We didn’t look in great detail at which were not found and why they were not.
- Get data out of WoS by using the Save to Other File Formats option. Specify the numbers of the records (we chose 1 through 138, i.e. all of them) (If you have lots, you may have to do this multiple times, as WoS has a limit of 500 records it will save in one go.). Select Record Content to be Full Record and Cited References, and File Format to be BibTex. Click Send and you should soon get a file
savedrecs.bib(it should appear in the folder where you downloads usually go). Rename the downloaded file to something meaningful, e.g. we named ours tofocal_corpus_from_WoS.bib.
Get information about the reference corpus
We got the reference corpus from WoS using the following search terms:
- ORGANIZATION-ENHANCED: (University of Zurich)
- WEB OF SCIENCE CATEGORIES: ( ENVIRONMENTAL SCIENCES OR REMOTE SENSING OR ECOLOGY OR PLANT SCIENCES OR GEOGRAPHY PHYSICAL OR ENVIRONMENTAL STUDIES )
- Timespan: 2013-2018.
- Indexes: SCI-EXPANDED, SSCI, A&HCI, CPCI-S, CPCI-SSH, BKCI-S, BKCI-SSH, ESCI, CCR-EXPANDED, IC.
This search returned over 2’000 papers that we saved the information from WoS about using the same approach as in step 6 in the previous section. We got five files of records for this reference corpus due to the 500 record save limit of WoS.
Load everything into R!
focal_corpus <- readFiles("~/Desktop/focal_corpus_from_WoS.bib") %>%
isibib2df() %>%
mutate(TC=as.numeric(TC),
Corpus="URPPGCB",
AUx=gsub(";","",AU),
num_authors=nchar(AU) - nchar(AUx))
D1 <- readFiles("~/Desktop/ref-corpus1.bib") %>%
isibib2df()
D2 <- readFiles("~/Desktop/ref-corpus2.bib") %>%
isibib2df()
D3 <- readFiles("~/Desktop/ref-corpus2.bib") %>%
isibib2df()
D4 <- readFiles("~/Desktop/ref-corpus2.bib") %>%
isibib2df()
D5 <- readFiles("~/Desktop/ref-corpus2.bib") %>%
isibib2df()
reference_corpus <- rbind(D1, D2, D3, D4, D5) %>%
mutate(TC=as.numeric(TC),
Corpus="Reference",
AUx=gsub(";","",AU),
num_authors=nchar(AU) - nchar(AUx))
rm(D1, D2, D3, D4, D5)
Make a co-authorship network (reference corpus only)
focal_corpus_biban <- biblioAnalysis(focal_corpus)
NetMatrix <- biblioNetwork(focal_corpus, analysis = "collaboration", network = "authors", sep = ";")
MM <- as(NetMatrix, "matrix")
MM <- MM[order(rownames(MM)), order(colnames(MM))]
diag(MM) <- 0
top_authors <- focal_corpus_biban$Authors[1:20]
these_cols <- colnames(MM) %in% names(top_authors)
these_rows <- rownames(MM) %in% names(top_authors)
top_MM <- MM[these_rows,these_cols]
set.seed(999)
chordDiagram(top_MM, symmetric = TRUE)
Combine and compare the focal and reference corpus
First combine into one object the data about the two corpi:
reference_corpus <- select(reference_corpus, intersect(names(reference_corpus), names(focal_corpus)))
focal_corpus <- select(focal_corpus, intersect(names(reference_corpus), names(focal_corpus)))
corpus <- rbind(focal_corpus, reference_corpus)
Get summary statistics about some characteristics of the articles, and graph them for comparison:
summ_stats <- group_by(corpus, Corpus) %>%
summarise(mean_cites=mean(TC),
sem_cites=sd(TC)/sqrt(n()),
mean_num_authors=mean(num_authors),
sem_num_authors=sd(num_authors)/sqrt(n()))
p1 <- ggplot(summ_stats) +
geom_point(mapping=aes(x=Corpus, y=mean_cites)) +
geom_errorbar(mapping=aes(x=Corpus,
ymin=mean_cites-sem_cites,
ymax=mean_cites+sem_cites),
width=0.1) +
ylab("Number of times cited")
p2 <- ggplot(summ_stats) +
geom_point(mapping=aes(x=Corpus, y=mean_num_authors)) +
geom_errorbar(mapping=aes(x=Corpus,
ymin=mean_num_authors-sem_num_authors,
ymax=mean_num_authors+sem_num_authors),
width=0.1) +
ylab("Number of authors")
p0 <- plot_grid(p2, p1)
ggsave(filename="~/Desktop/corpus_comparison.jpg", plot=p0, width = 6, height = 4)